Data Anonymization Techniques: Tokenization and Bloom Filters
Understanding tokenization for matching data
Organizations across healthcare need to leverage de-identified real-world data (RWD) for single research projects as well as enterprise data initiatives. This requires bringing together disparate datasets at the patient level.
HIPAA has a list of 18 pieces of identifying information that must be removed in order to satisfy the Safe Harbor method of de-identification, and offers further guidance on how parts of patient data can be included in a record while still qualifying as “de-identified.” The goal, of course, is to protect patient privacy, but unfortunately, if a patient’s identifying information is completely removed from a record, there is no way to combine a patient’s healthcare data in one record (e.g., a hospital stay) with data in another record (e.g., a pharmacy prescription after being discharged from the hospital).
To match patient data across datasets, Datavant Connect provides tokenization software to de-identify patient health information (PHI) and subsequently relink it with other datasets while maintaining the privacy of personally-identifiable information (PII). Tokenization software replaces PII with an encrypted “token” that can’t be reverse-engineered to reveal the original information. With this technique, we can create the same patient-specific tokens in any dataset, which means two different datasets can be combined using the patient tokens to match corresponding records while also maintaining the privacy of the PII in those records.
Our match process makes a ternary agreement vector from a pair of patient records: “true” if records match, “false” if they don’t match, or “null,” if they cannot match because you can’t compare them, when, for example, one record is missing data in a particular field of comparison, like a middle name:
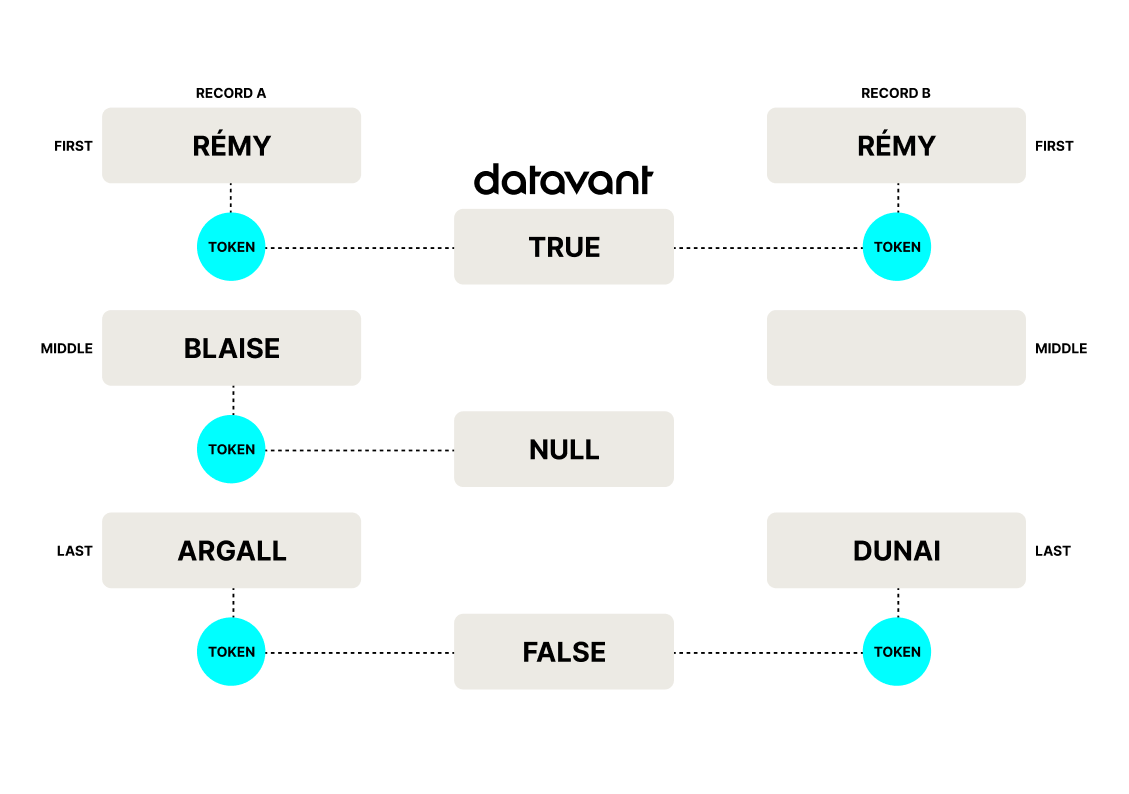
This ternary vector feeds into a machine learning model, which then determines if the records being compared belong to the same patient or not. You can read a more in-depth summary of our tokenization process here.
One potential alternative strategy for matching data across datasets is through the use of Bloom filters.
What’s a Bloom Filter?
Conceived by Burton Howard Bloom in 1970, a Bloom filter is a probabilistic data structure used to determine whether a particular element is a member of a given set. A query will not tell you with total certainty that an element is in a set, but will instead give you the likelihood that an element is in a set, or will tell you that it is definitively not in the set. Depending on your tolerance levels, you may receive false positives, but you will never receive a false negative. For more in-depth tutorials on Bloom filters, there are some excellent resources available at Geeks for Geeks coding introductions, Github, and even Wikipedia.
(Incidentally, fruit flies have been shown to exhibit a neural data structure for novelty detection that functions similarly to a Bloom filter.)
When the available data complicates attempts at matching
Bloom filters could be used to encode PII into numerical vectors that can be easily compared. The main advantage of Bloom filters over tokenization is that they are more robust in the face of minor discrepancies of data, like clerical errors, which can complicate matching records with precision.
To illustrate this, consider this record from a patient hospital visit with just a few PII elements:
JOHN, O’SHEA, MALE, 10/21/1967, 33756
At a repeat hospital visit, a clerical error occurs while Mr. O’Shea’s data is entered in the system, and a new record is created with just a single misspelling:
JOHN, O’SHEE, MALE, 10/21/1967, 33756
Because Datavant’s tokenization relies heavily on full last names for matching, there is a chance that these records may not be matched as belonging to one individual because of the typo.
Again, within the matching process generally, there are two primary concerns: accuracy of record match, and ability to maintain privacy. Let’s discuss matching first.
Probabilistic matching via Bloom filters
Bloom filters provide partial matching that addresses minor discrepancies, like typos. They rely on hashing to create a filter that is itself a binary vector of some given length L alongside a set of k hash functions háµ¢ that map data elements into the integer range 1…L.
For example, to encode the data element “JOHN” into a Bloom filter, you could hash each letter of the name using each of the hash functions háµ¢ and set the bits at those positions to 1.
Here is an illustration of how this works:
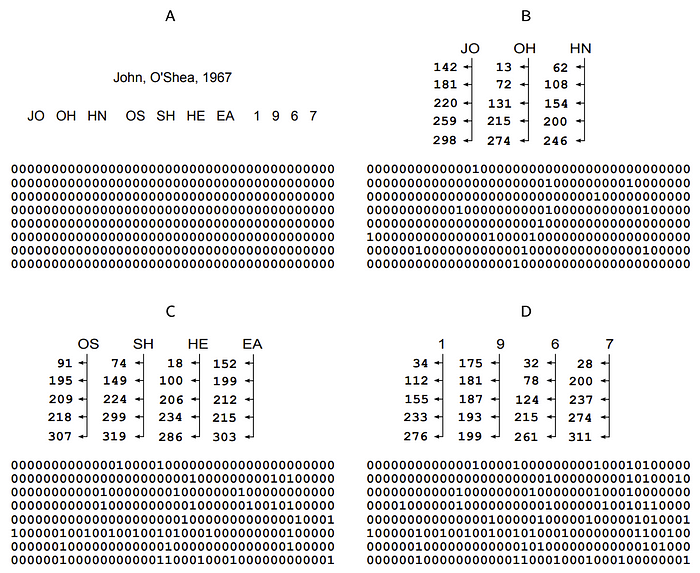
Here, a record with three of the five PII elements given above is hashed into a large Bloom filter using five hash functions (the vertical columns of numbers beneath each sub-element). What the numbers under each sub-element (bigrams for pairs of letters or unigrams for single date numbers) represent are the positions in the Bloom filter that are set to 1 on each pass.
Once a Bloom filter is populated for a record, it can be matched against Bloom filter encodings of other records by computing a distance between the two binary vectors. The most common distances to use are the Hamming distance (the minimum number of substitutions required to change one string into the other) defined here as the number of exact bit matches, and the DICE index (a statistic used to gauge the similarity of two samples), which underweights matches on bit values of 0. We can write this out as an equation:
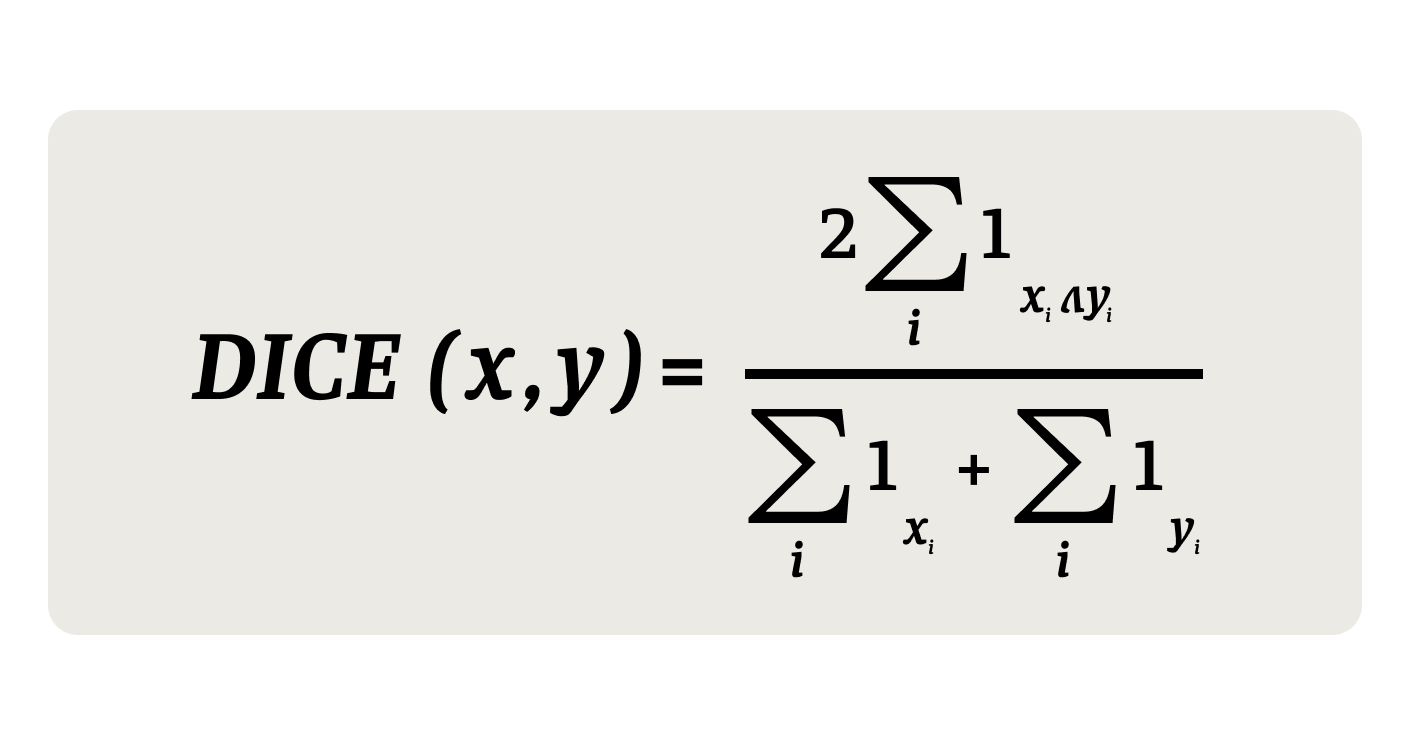
Exploring how we might use Bloom filters
We experimented with a model trained using Bloom filters to use pairwise record comparison vectors that included the following profile fields:
[DICE Index, Gender Feature, ZIP3 Feature, YOB Feature, Collision Score]
ZIP3 is what Datavant uses instead of full zip codes to define “zip areas” (based on HIPAA Safe Harbor rules), and YOB is year of birth, which we use instead of full birthdate. The Collision Score is generated based on the relative frequency of combinations of first name / last name / decade of birth. Every year, several thousand people are born with the same first name-last name combinations in the U.S. The Collision Score helps us determine the likelihood that records sharing the same name/birth year combination belong to the same person.
Instead of generating outcomes of “match” / “no match” / “I don’t know,” as we do in our tokenization process, the Bloom filter gives a number between 0 and 1, which is the filter confidence score that these two records represent a match, or “I don’t know” if it’s impossible to compare a record. (Again, e.g., if one record has a middle name and the other record does not, then it’s impossible to compare middle names).
In this experiment, Bloom filters were only used to compute the DICE index. Bloom filters are often very large binary vectors and classical machine learning models struggle with data that is too high-dimensional. Because the DICE index captures small differences between records, it mitigates the clerical error problem shown in the first two examples above. The DICE index between
JOHN, O’SHEA, MALE, 10/21/1992, 33756
JOHN, O’SHEE, MALE, 10/21/1992, 33756
is 0.96 (almost identical), while the DICE index between
JOHN, O’SHEA, MALE, 10/21/1992, 33756
JANE, DOE, FEMALE, 09/12/1964, 01111
is 0.20 (little overlap).
While we cannot confirm with absolute certainty a match in the first example, we can say that it is very likely that these two records belong to the same individual. In the second example, the very low degree of overlap tells us that these records certainly do not belong to the same individual.
Comparing the effectiveness of Datavant Connect tokenization with Bloom filters in record matching
To evaluate Bloom filters against our tokenized model, we used two samples for training and evaluation:
- A small sample of 300,000 real world data pairs that we had previously identified through a pre-filtering process as potentially matching records (i.e.: where at least one token matches);
- A large sample of approximately 3 million pairs that we had previously identified as potentially matching records.
The datasets were split into an 80% training sample and a 20% testing sample. For the sake of the comparison, we did not perform hyper-parameter tuning or cross-validation.
For each dataset we trained two models:
- Our production token comparison model that used a small set of all potential tokens, which are representative of the data a customer has available to tokenize alongside the fields of [Gender, Zip3, YOB, Collision Scores];
- A model that used only the Bloom filter feature alongside profile fields and Collision Scores.
Results on Small and Large Datasets
To evaluate performance, we looked at three metrics:
AUC-PR, Recall@95%, and Recall@99%.
AUC-PR (area under curve-precision recall) is a performance metric that is useful for working with imbalanced data in a setting where the goal is to locate positive examples. In our case, we are looking for positive matches between profile fields across medical records, and we do not want false positives, i.e.: we do not want to return a “match” between records that do not belong to the same person. The percentage given in the table below is calculated by dividing # positive examples / total # examples.
The other performance metric we considered is Recall@X%. Whereas AUC-PR attempts to identify what proportion of positively identified matches was correct, Recall@X% attempts to identify what proportion of all positive matches were correctly identified, and is also measured in percentage points. You can read more about precision recall as it relates to machine learning here.
Internally, we use Recall@95 to evaluate new models. Recall@95 is the recall that a model achieves at a fixed 95% precision rate. In the table below, tokens achieved 90% recall when the model threshold was set to 95% precision rate. We use this metric because both we and our clients often require this as a minimum level of precision for matching to be considered successful. Favoring higher precision in recall reduces the likelihood of returning false positives. Optimization of models then becomes a matter of maximizing recall for that minimum level of precision.
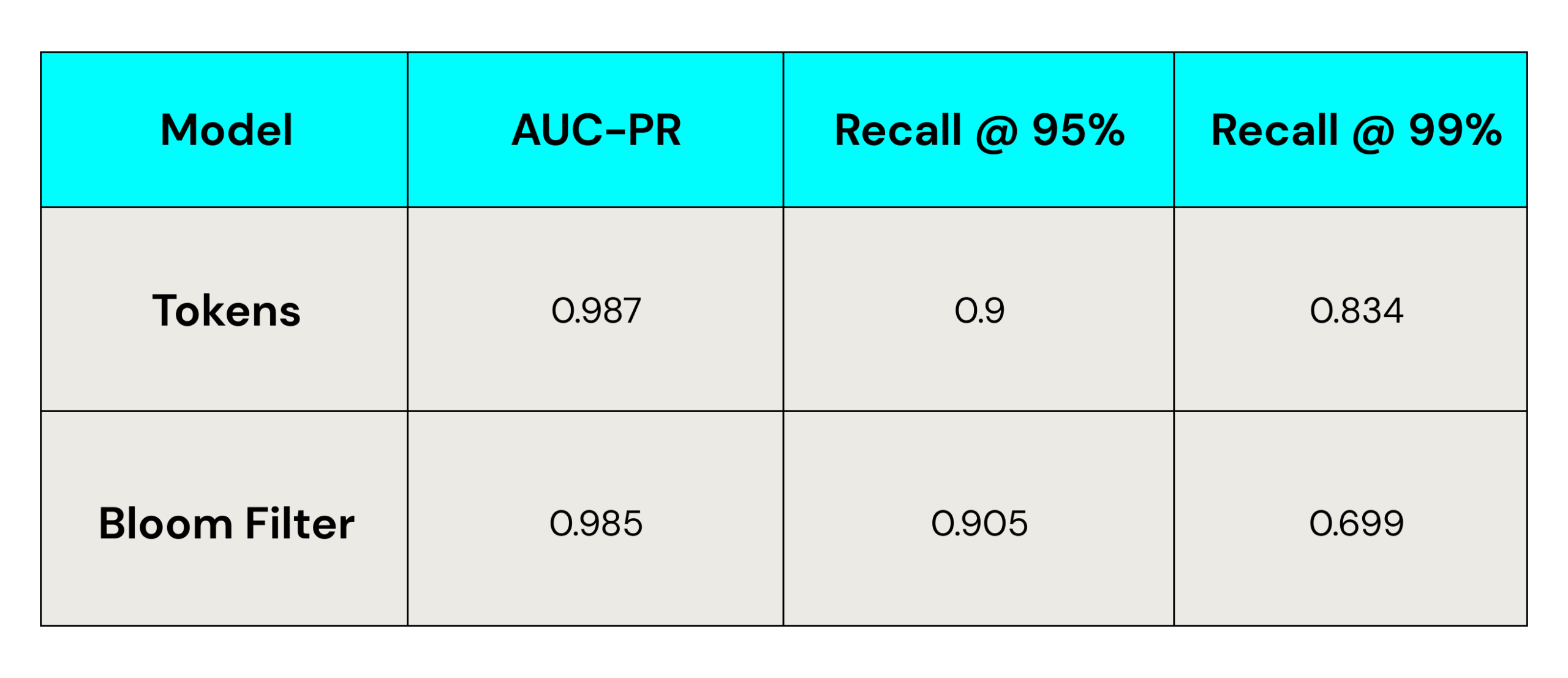
Results on Small Dataset
Results on Large Dataset
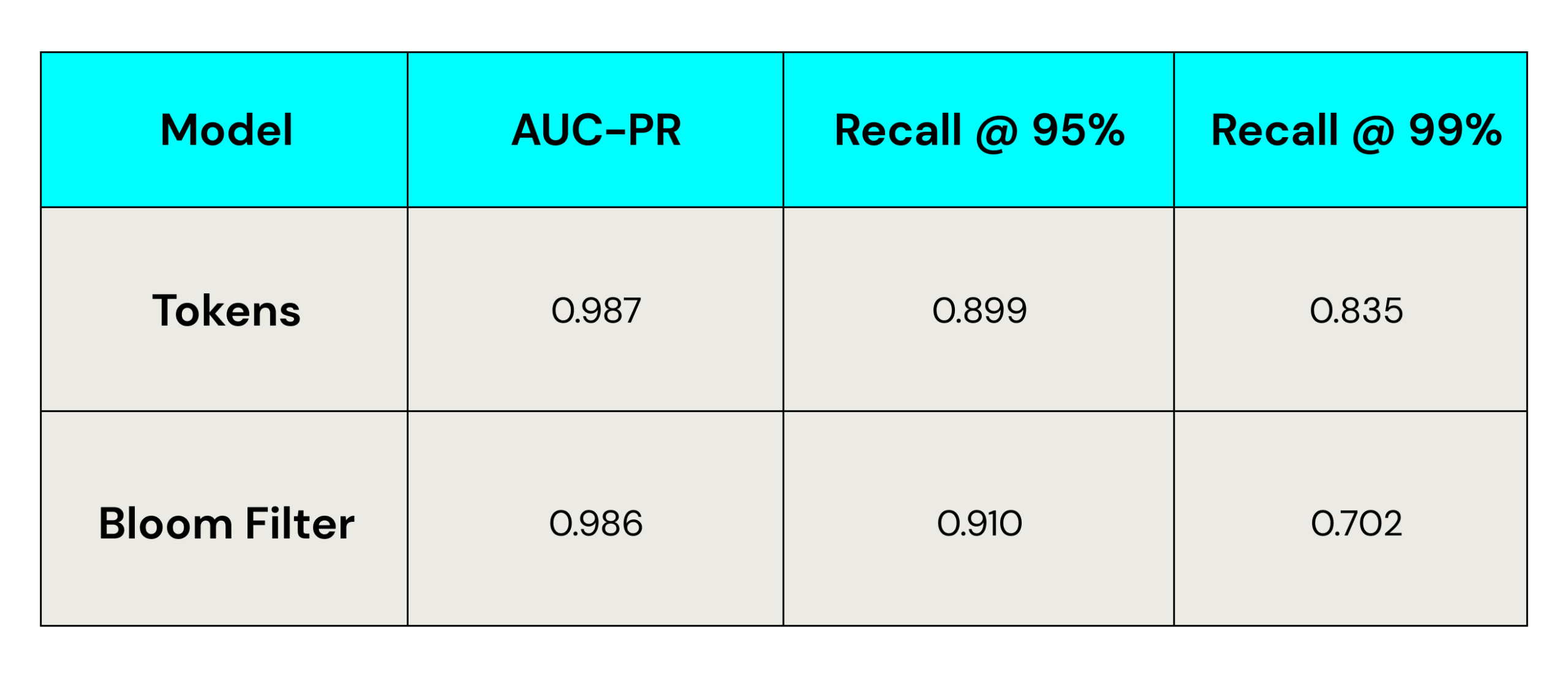
You can see in the above charts that in both datasets, tokens have a slight edge over Bloom filters when evaluated using AUC-PR, and Bloom filter models have a slight edge over tokens when evaluated with Recall@95%. However, Bloom filters had a significant drop in successful matching at Recall@99% precision (correctly identifying 99% of matches).
Where Bloom filters shine
One situation where Bloom filters outperform tokens is in the presence of significant clerical errors on token-critical PII. For example, if there is a high likelihood of last name misspellings, we determined that a Bloom filter approach will likely perform better in determining matches. However, the number of clerical errors in the dataset used for this comparison was small compared to other errors, so the performance bump offered by Bloom filters was negligible here.
Prefiltering: a disadvantage in the Bloom filter approach to matching
Datavant relies on tokens to quickly filter the list of potential matches for any given record. We tokenize each profile field within a record (First name, Last name, ZIP3, etc.), so our assumption that two records are unlikely to match unless at least one of the tokens matches across records means we can do a quick and easy-to-parallelize pre-filtering step when we receive new records.
In certain circumstances, Bloom filters allow for a technique called blocking (performing a subset comparison to reduce the total number of necessary comparisons), but the pre-filtering strategy employed by our tokenization technique is more efficient at this. Efficiency at this stage helps improve performance downstream by eliminating the need to compare each new record with every other record to locate potential matches.
There could be an alternative workaround for prefiltering with Bloom filters. Remember that one of our means for comparing Bloom filter encodings is via Hamming distance, the minimum number of substitutions required to change one string into another. In such spaces, there are efficient approximate algorithms for retrieving the closest 100 (or any other n) neighbors of a given point. A benchmark of the most popular algorithms is available here.
Matching would then involve two steps. Given a new record, we would:
- Find the closest 100 already-seen records using an approximate nearest neighbor algorithm;
- Do a full comparison on those 100 records to create new match links.
In this procedure we could miss true positive matches, but we would attempt to mitigate that problem by allowing for transitive connections (i.e.: A matches B, B matches C, therefore A matches to C through transitive connection).
Preserving privacy: the mission-critical performance metric
The other key metric of the effectiveness of the de-identification and match procedure is its potential for preserving the privacy of PII. Datavant’s tokenization process is secure, transparent, and auditable.
We see Bloom filters as vulnerable to Frequency attacks. A malicious user could re-identify parts of the dataset by looking at, for example, the frequency of set bits in a location of the Bloom filter.
One possible guard against this would be to replace the filter itself. The principal feature of the binary vectors generated in the Bloom filtering process that we care about is the distance between the two binary vectors being used to find matches. If instead of saving the binary vector, we save the output of a map:
f:[0,1]ᴸ → R⿠such that

then we could potentially dodge frequency attacks.
As mentioned at the very top of the post, Datavant is committed to preserving privacy throughout the data exchange process. In order for us to consider using Bloom filters in production, they would need to overcome the extremely significant hurdle of independent auditability in this mission-critical area of performance.
Takeaways
- Bloom filters offer a marginal advantage over Datavant’s tokenization in situations where there are likely to be many small discrepancies between matching records. This marginally better performance they provide at Recall@95 is outweighed by:
- their vulnerability to Frequency attacks,
- lower performance at Recall@99, and
- current lack of auditability.*
- Finally, we believe that they are also harder to fine-tune for performance than the machine learning models involved in our current tokenization.
*This is not to say that Bloom filters could not become secure, or could not be audited, but that such an audit would require extensive time and resources.
N.B. These conclusions are drawn from evaluating on currently available datasets and may change with new data.
Authored by Sebastian Claici and Nicholas DeMaison, with significant input from Jonah Leshin, Varun Lahoti, Arjun Sanghvi, Carlos Guzman, Aneesh Kulkarni, and Bob Borek.
About the Authors
Sebastian Claici holds a PhD in Computer Science from MIT, where he was also a research assistant. He joined Datavant as a Data Scientist in 2021. Connect with Sebastian via Linkedin.
Nicholas DeMaison enjoys telling simple stories about complex things. Connect with Nick via LinkedIn.
Considering joining the Datavant team? Check out our careers page and see us listed on the 2022 Forbes top startup employers in America. We’re currently hiring remotely across teams and would love to speak with any new potential Datvanters who are nice, smart, and get things done and want to build the future tools for securely connecting health data and improving patient outcomes.



